MuLTEE projects
As we continue the evolution of the MuLTEE, we are examining a range of projects using this model system.
Evolution of cellular differentiation
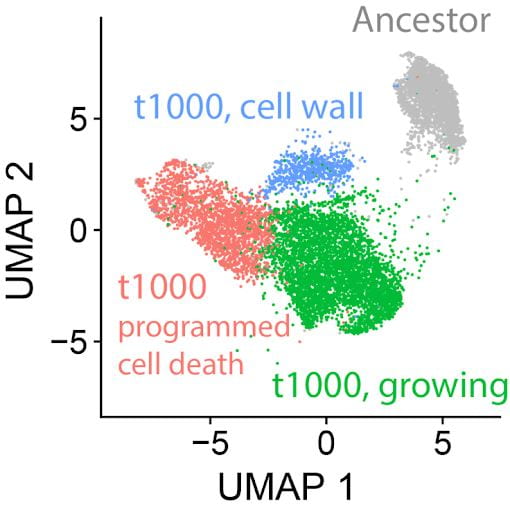
Cellular differentiation is arguably the single greatest benefit available to multicellular organisms. To explore how differentiation arises de novo, we have been using single-cell RNA-seq to examine cell-level gene expression over 1000 transfers (~5,000 generations). We see the emergence of three putative cell types evolving (see UMAP plot), which each are relatively specialized on cellular growth, cell wall biogenesis, and programmed cell death. We are currently investigating their spatial localization, biological function, developmental dynamics, and genetic regulation.
Population genomics
We are currently the genomes and transcriptomes of snowflake yeast across the first 1000 transfers of the MuLTEE, examining how differences in metabolism and population size affect multicellular evolution. Specifically, as larger organisms evolve, their population size reciprocally plummets, as the total number of cells per population is relatively constant. Following the pioneering work of Michael Lynch, we’re examining how the evolution of multicellularity impacts population size and thus the relative efficacy of purifying selection, and the impact of this on structural variation/genome size.
Epigenetic mechanisms of multicellular adaptation
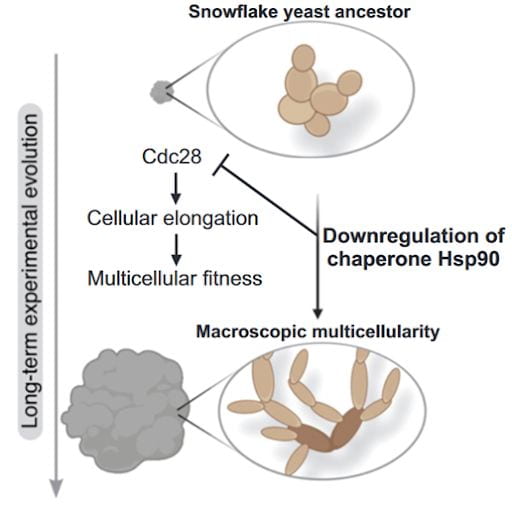
In addition to examining genetic contributors to multicellular evolution, our group is examining epigenetic mechanisms, specifically the role of downregulation of the core chaperone protein HSP90. This chaperone protein is convergently downregulated during the MuLTEE, which contributes to the origin of novel multicellular traits by changing the activity of client proteins. We’re very interested in both the short-term and long-term impacts of altered proteostasis.
The emergence of biophysical toughness
We are interested in how early multicellular organisms evolve to be biophysically tough, able to weather new (and large) forces that threaten to destroy the organism. One of the ways that branching, growing organisms can do this is by entangling their branches, wrapping around each other, and holding on to distribute stresses. Having seen entanglement evolve convergently in the MuLTEE, we’re exploring how this behavior arises in living systems more broadly.
Long-range fluid flows
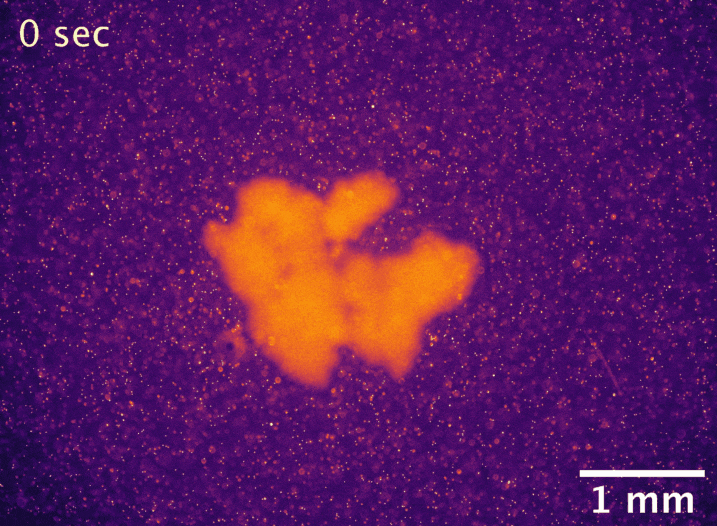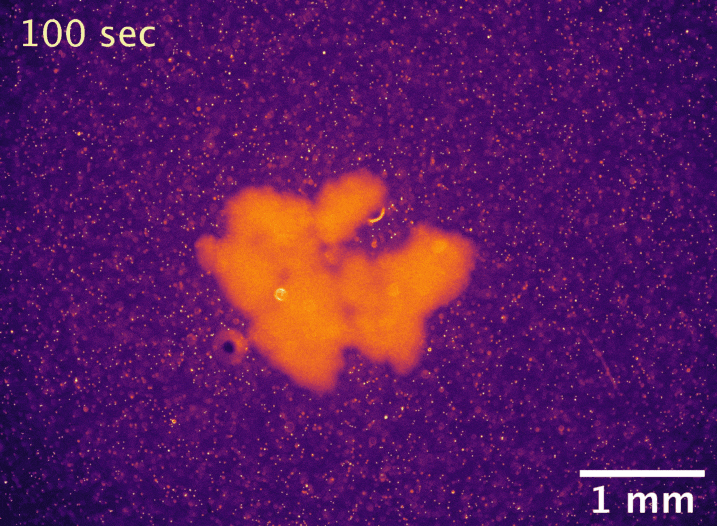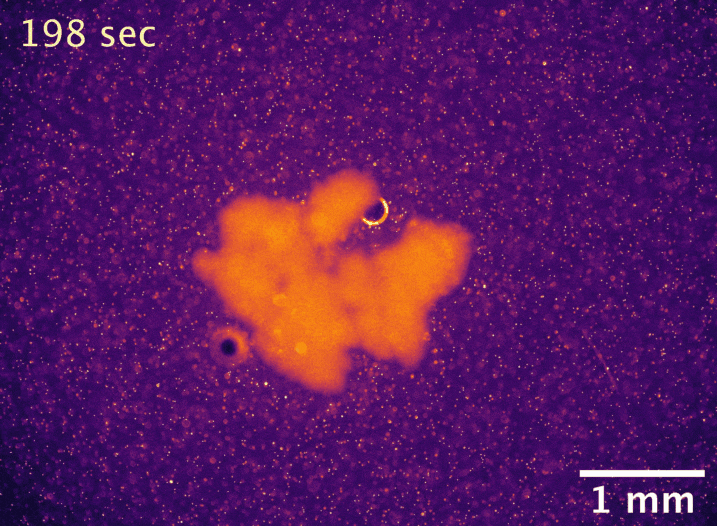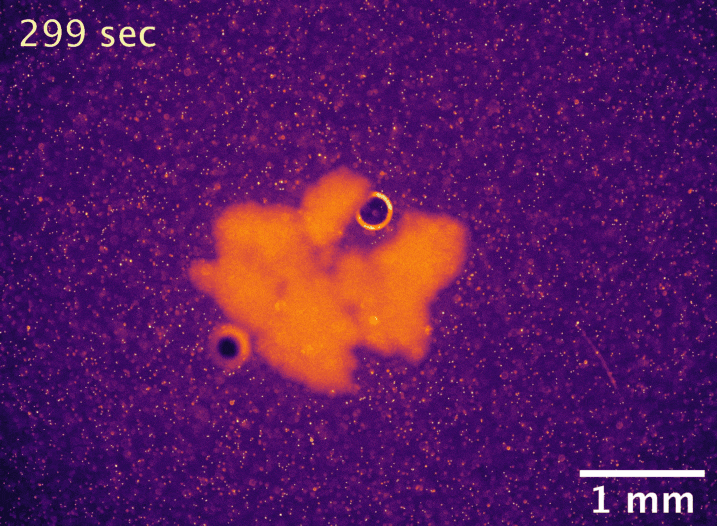
Usually, the larger an undifferentiated group of cells grows, the more problems it has with diffusion limitation. The cells on the outside take up all the nutrients first, starving the inside cells and setting a microscopic upper bound on size. However, macroscopic snowflake yeast, despite having no cilia or circulatory system, are able to grow to centimeter-size scales in still media with no reduction in their growth rate. How can they do this? In collaboration with Shashi Thutupalli’s group at NCBS, we found that snowflake yeast clusters’ own metabolism drives fluid flows with speeds of 5-10µm/s in liquid media, and we believe these flows help nutrients penetrate through the clusters. Using micron-sized fluorescent beads, you can see the flows below. We hypothesize that the flows are created due to variation in fluid density caused by gradients of nutrients and waste products. These fluid flows represent another way that early multicellular organisms might have relied on emergent phenomena to overcome the biophysical limits imposed by large size.
Entrenchment of multicellularity
The transition to multicellularity has been shown to occur readily, but reversion back to unicellularity can be favored if there is no longer a selection for the group. However, some multicellular lineages appear to be evolutionary stable, as there are no living unicellular organisms descended from animals or plants. This raises the question of what makes multicellularity stable. And also how the fitness of a cell changes over evolutionary time in a newly evolved multicellular organism, as the evolved phenotypes that benefit the group may not benefit the individual. To explore these questions, we are using various approaches. First, we are using engineered unicellular revertants of the evolved strains in the MuLTEE. Second, we are measuring the fitness of cells in a multicellular cluster. Third, we are conducting directed evolution experiments to evolve unicellularity from the evolved strains of the MuLTEE. Finally, we are using digital “organisms” to study this phenomenon (preprint)


Divergence and coexistence
The evolution of multicellular life spurred evolutionary radiations, fundamentally changing many of Earth’s ecosystems. Yet little is known about how early steps in the evolution of multicellularity transform eco-evolutionary dynamics, e.g., via niche expansion processes that may facilitate coexistence. Using the MuLTEE, we examine how the evolution of multicellularity can drive niche partitioning. In the obligately aerobic treatment of the MuLTEE, we observe the emergence of two distinct, specialized lineages from a single multicellular ancestor: a small genotype that is specialized on growth, and a large phenotype specializing on survival during settling selection. These two lineages last shared a common ancestor at the start of our experiment, and coexistence is maintained over thousands of generations. Through modeling and experimentation, we demonstrate that this coexistence is maintained by a trade-off between organismal size and competitiveness for dissolved oxygen, a key resource in this treatment (preprint).

Other projects
The MuLTEE is our lab’s flagship project, but we have a number of other projects going exploring the evolution of multicellularity and microbial social evolution.
Synthetic Biology
Yeast are a great model organism for genetic manipulation. We can recapitulate traits that evolved in the MuLTEE, for instance testing biophysical hypotheses for how changes in cell-level traits affect the emergence of multicellular traits. We also use synthetic genes to endow yeast with novel traits that are relevant to the evolution of multicellular organisms, but which do not naturally arise in our experiment. For instance, we can engineer yeast’s oxygen metabolism, or affect O2 uptake rates by providing them with genes encoding O2-binding proteins (globins and myohemerythrins). We can even engineer yeast to utilize light energy with microbial rhodopsin genes (preprint). We examine how these traits influence multicellular evolution, testing basic hypotheses about ecological and organismal factors influencing the evolution of complex life.
Multicellular life cycles
We have several projects underway which use synthetic biology to modify the multicellular life cycle of a yeast genotype (e.g., modifying clonal vs. aggregative development, or facultative vs. obligate multicellularity), then examine how these life cycle traits affect subsequent multicellular evolution. In combination with mathematical modeling, this approach allows us to determine how relatively small differences in the mode of multicellular group formation can have large impacts on the subsequent evolution of novel multicellular traits.
Microbial Sociality
In a long-term collaboration with Brian Hammer’s group, we explore how microbial antagonism, mediated by the T6SS, shapes interactions within and between species, and how these can affect the spatial dynamics of microbial populations and subsequent evolution of cooperation/conflict. Ongoing projects combine experiments and theory to examine the costs of T6SS expression (preprint). We are also using experimental evolution to study how the target bacteria evolve to resist T6SS attack (preprint).